The objective is to create an advanced Content Delivery Network (CDN) supporting medical and research systems that make extensive use of genomic data sets. ARES will implement a pilot project in order to gain a more detailed understanding of network problems relating to a sustainable increase in the use of genome data sets for diagnostic purposes. Suitable management policies for very large sets of big files will be identified, in terms of efficiency, resiliency, scalability, and QoS in a distributed CDN environment. In addition, ARES will make available, for the GÉANT network, suitably designed tools for deploying any CDN services handling large data sets, beyond genomes.
The increasing scientific and societal needs of using genomic data and the parallel development in sequencing technology has made affordable the human genome sequencing on large scale. In the next few years, a lot of applicative and societal fields, including academia, business, and public health, will require an intensive use of the massive information stored in the DNA sequence of the genome of any living body. Access to this information, a typical big data problem, requires redesigning procedures used in sectors such as biology, medicine, food industry, information and communication technology (ICT), and others.
The strategic objective of the project ARES (Advanced networking for the EU genomic RESearch) is to create a novel Content Distribution Network (CDN) architecture supporting medical and research activities making a large use of genomic data.
The main research objectives of ARES are:
- To gain a deep understanding of network problems relating to a sustainable increase in the use of genomes and relevant annotations for diagnostic and research purposes.
- To allow extensive use of genomes through the collection of relevant information available on the network for diagnostic purposes.
- To identify suitable management policies of genomes and annotations, in terms of efficiency, resiliency, scalability, and QoS in a distributed environment using a multi-user CDN approach.
- To make available the achieved results to extend the service portfolio of the GéANT network with advanced CDN services.
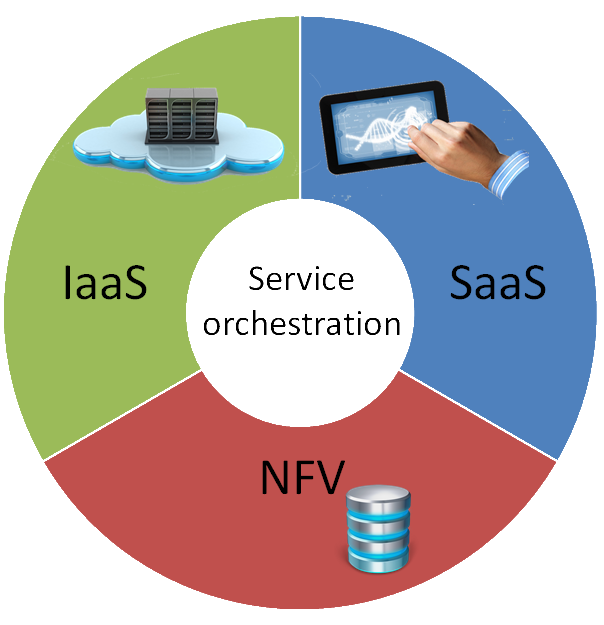
The technological pillars over which the ARES project rely are :
- infrastructure as a service (IaaS) for managing processing of genomic files in data centers with virtual machines (VMs), managed through OpenStack tenants;
- software as a service (SaaS) for managing the access of end users to the processing platform (web interface);
- NFV caching modules in routers and in data center VMs, implemented through the NetServ platform;
- a central orchestration engine, which uses the NSIS signaling to retrieve the status of computing and caching resources.
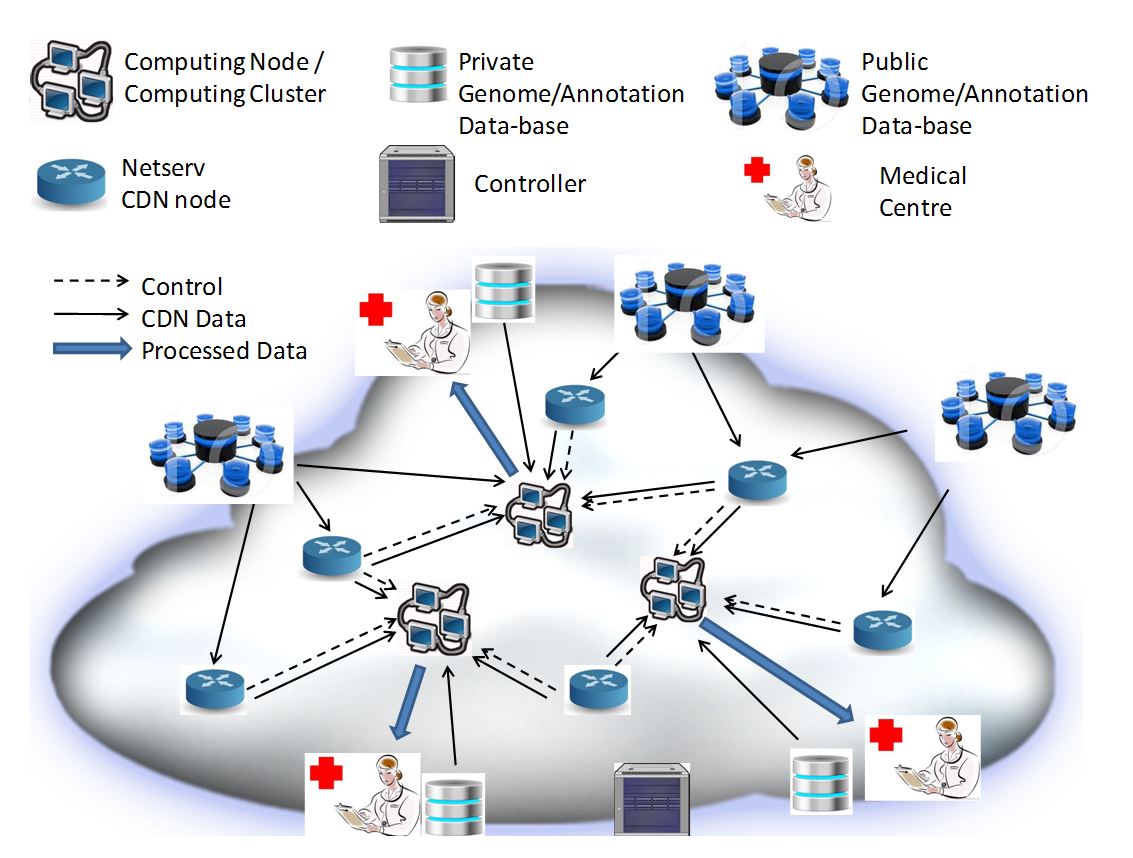
Steps of the experiments:
- A Medical Centre asks for the genomic analysis service. The service determines the genomic annotation files required to execute the processing, which consists of searching for the already known sequences included in genome annotations for potential changes (e.g. mutations) and similar variations.
- The service request is done by the Medical Centre though a web-based cloud interface. It triggers on the NetServ Genomic CDN Manager (GCM) a resource discovery procedure based on NSIS signaling on NetServ nodes, which identifies, by solving an associated optimization problem, the set of one or more candidate nodes for executing the genome processing software.
- The NetServ GCM triggers a service for transferring the genome processing software over the selected Computing Node/Cluster using the NetServ ActiveCDN service.
- The local NetServ Controller bundle triggers the software modules. They are now ready to receive input data.
- The Medical Centre sends the patient’s genome data to the Computing Node/Cluster.
- The NetServ ActiveCDN service downloads the needed genomic information files from the Public Genome/Annotation data-bases into the Computing Node/Cluster.
- The local NetServ Controller triggers the service execution over the Computing Node/Cluster.
- The parameters estimates obtained from data processing are returned to the Medical Centre, through the web-based cloud interface.
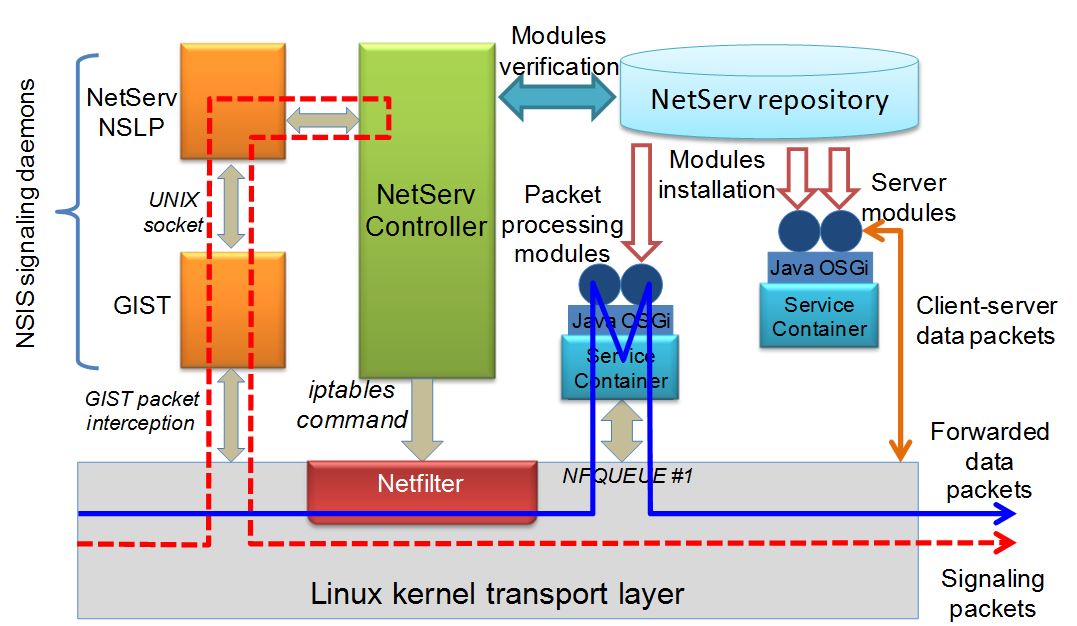
The following picture depicts the developed service virtualization architecture. The applications use building blocks to drive all network operations, with the exception of packet transport, which is IP-based. The resulting node architecture, designed for deploying in-network services, is suited for any type of nodes, such as routers, servers, set-top boxes, and user equipment. This architecture is targeted for in-network virtualized service containers and a common execution environment for both packet processing network services and traditional addressable services (e.g. a Web server). It is thus able to eliminate the mentioned dichotomy in service deployment over the Internet and administrators can be provided with a suitable flexibility to optimize resource exploitation. It is currently based on the Linux operating system, and can be installed on the native system or inside a virtual machine. It includes an NSIS-based signaling protocol, used for dynamic discovery of nodes hosting the NetServ service environment, and service modules deployment therein. NSIS is an IETF standard consisting of two layers, NSIS transport layer protocol (NTLP) and NSIS signaling layer protocol (NSLP); the latter contains the specific signaling application logic.
GIST (General Internet Signaling Transport protocol), is a widely used implementation of NTLP. GIST uses existing transport and security protocols to transfer signaling (i.e. NSLP) messages on behalf of the served upper layer signaling applications.
It provides a set of easy-to-use basic capabilities, including node discovery and message transport and routing. Since its original definition, GIST allows transporting signaling message according to two routing paradigms. It provides end-to-end signaling, that allows sending signaling messages towards an explicit destination, and path-coupled signaling, that allows installing states in all the NSIS peers that lie on the path between two signaling peers.
Our implementation of a third routing paradigm, named off-path signaling, that allows sending signaling message to arbitrary sets of peers, totally decoupled from any user data flow. A peer set is called off-path domain. Thanks to the extensibility of GIST, other off-path domains can be implemented as they become necessary for different signaling scenarios.
The current implementation of the NetServ signaling daemons is based on an extended version of NSIS-ka, an open source NSIS implementation by the Karlsruhe Institute of Technology. As mentioned above, NSIS wraps the application-specific signaling logic in a separate layer, the NSLP. The NetServ NSLP is the NetServ-specific implementation of NSLP. The NetServ NSLP is able to manage the hot-deployment of bundles on remote nodes.
The NetServ controller coordinates NSIS signaling daemons, service containers, and the node transport layer. It receives control commands from the NSIS signaling daemons, which may trigger installation, reconfiguration, or removal of both application modules within service containers and filtering rules in the data plane by using the netfilter library through the iptables tool. Each deployed module has a lifetime associated with it. It needs to be refreshed by a specific signaling exchange by its lifetime expiration, otherwise it is automatically removed. The NetServ controller is also in charge of setting up and tearing down service containers, authenticating users, fetching and isolating modules, and managing service policies.
Service containers are user-space processes. Each container includes a Java Virtual Machine (JVM), executing the OSGi framework for hosting service modules. Each container may handle different service modules, which are OSGi-compliant Java archive files, referred to as bundles. The OSGi framework allows for hot-deployment of bundles. Hence, the NetServ controller may install modules in service containers, or remove them, at runtime, without requiring JVM reboot. Each container holds a number of preinstalled modules, which implement essential services. They include system modules, library modules, and wrappers of native system functions. The current prototype uses Eclipse Equinox OSGi framework.
The NetServ repository, introduced in the NetServ architecture for management purposes, includes a pool of modules (either building blocks or applications) deployable through NetServ signaling in the NetServ nodes present in the managed network.
We will address these following specific research challenges:
- definition of the service requirements for genome and annotation exchange;
- deployment of a suitable architectural framework, for modular and virtualized services, able to provide suitable insights for the service operation and relevant perception in a research environment;
- deployment of mechanisms and protocols for service description, discovery, distribution, and composition.
- experimental performance evaluation, in terms of QoE, scalability, total resource utilization, signaling load, performance improvement and coexistence with legacy solutions, dynamic deployment and hot re-configurability.
Advanced CDN and virtualization services
Our experiments aims to investigate (i) which network metric is suitable for the successful and efficient usage of genomes and annotations for both diagnosis and research purposes, and for associating requesting users with an available cache (such as total number of hops, estimated latency, queue length in servers, available bandwidth), (ii) the best policy for dynamically instantiating and removing mirrors, with reference to the user perception, the signaling load, and the information refresh protocols, (iii) the achievable performance when only a subset of nodes, with a variable percentage, execute the NetServ modules analyzed. In this latter case, our enriched off-path signaling of the NSIS modules, mentioned in the previous section will be essential, since it allows discovering the nodes actually executing NetServ and available services therein.
In the proposed scenario we assume that each network point of presence (PoP, see Figure 1) has a certain amount of storage space and computational capabilities hosted in one or more servers and managed through a virtualization solution. In this virtualized infrastructure, we assume that a specific node executes a virtual machine that hosts NetServ, and that the virtualized resources of the PoP are managed by OpenStack. OpenStack is a cloud management system that allows using both restful interfaces to control and manage the execution of virtual machines (VMs) on the server pool. It allows retrieving information about the computational and storage capabilities of both the cloud and the available VMs. It also allows triggering the boot of a selected VM with a specific resource configuration. Thus, it is possible to wrap, inside a certain number of VMs, different existing software packages able to process genome files. The NSIS signaling indicates the required computational capabilities for a specific processing service, which is mapped into the resource configuration of the VM that executes the service. Virtual machine image files are stored within the storage units of the PoP managed by OpenStack and also available for download through a restful interface. In fact, OpenStack allows also to automatically inject, in the managed server pool, new VMs by specifying an URL which points to the desired VM image, thus allowing the system to retrieve the relevant files from other locations (i.e. other GéANT PoPs).
An ActiveCDN bundle can either be executed on the same NetServ container hosting the OpenStack Manager service module, or on an instance of NetServ deployed within a different VM. The ActiveCDN module is in charge of managing the restful interface for distributing both the genomic data files and the VM image files. It implements the HTTP server function by using the NetServ native support for Jetty, a light-weighted HTTP server implemented in Java, which allows creating restful interfaces and managing different servlets.
Finally, at least one of the NetServ nodes will run the decision system, implemented in the GCM NetServ bundle. The GCM node will make an intensive use of the NSIS capabilities. In the proposed architecture, the GCM can use a Hose off-path signaling scheme, which extends off-path signaling to the NSIS-enabled nodes adjacent to those along the IP path between the signaling initiator and the receiver. The NetServ GCM can be replicated in multiple locations for scaling out the service.



